Dear dask community,
We are working on using dask for image processing of OME-Zarr files. It’s been very cool to see what’s possible with dask. Initially, we mostly did processing using the mapblocks API and things were running smoothly. But recently, we’ve started to handle some more complex cases that mapblocks doesn’t seem to handle (as far as I understand) and where we’re relying on dask.delayed & indexing. The core motivation for this is that we need to process lists of user-specified regions of interests in a large array and those regions do not necessarily have uniform sizes & positions in the array or map back to the chunks of our Zarr file or our dask array.
As an example, we may want to do something like this:
We have a list of regions of interest in a huge array, only process these small regions. If we have a dask array of 500x100kx100k, but we’re only interested in 50 cubes of 100x1000x1000 at random positions. How do we best process this? The best way we’ve found so far is using delayed functions & indexing of the dask array.
As an initial implementation of this new, more flexible approach, we’ve tried to implement a simple case that could still be done with mapblocks (see simplified code examples below). While this works, it seems to introduce slower running code and much more memory-inefficient processing. Is our approach (described below) something that should be supported and we’re doing it wrong? Or is there a better way to achieve the same goal? Does anyone have tips in how we make this more memory efficient?
Details:
What are we trying to achieve? Process a Zarr file by applying some functions to chunks of the data and saving it back into a new Zarr file.
We created a simplified, synthetic test example of that.
Code to generate synthetic Zarr test data:
import numpy as np
import dask.array as da
shapes = [
(2, 2, 16000, 16000),
(2, 4, 16000, 16000),
(2, 8, 16000, 16000),
(2, 16, 16000, 16000),
]
for shape in shapes:
shape_id = f"{shape}".replace(",", "").replace(" ", "_")[1:-1]
x = da.random.randint(0, 2 ** 16 - 1,
shape,
chunks=(1, 1, 2000, 2000),
dtype=np.uint16)
x.to_zarr(f"data_{shape_id}.zarr")
Code to process the data using a mapblocks approach
SIZE = 2000
# Function which increases value of each index in
# a SIZExSIZE image by one (=> pseudo-processing)
def shift_img(img):
return img.copy() + 1
def process_zarr_mapblocks(input_zarr):
out_zarr = f"out_{input_zarr}"
data_old = da.from_zarr(input_zarr)
n_c, n_z, n_y, n_x = data_old.shape[:]
dtype = np.uint16
print(f"Input file: {input_zarr}")
print(f"Output file: {out_zarr}")
print("Array shape:", data_old.shape)
print("Array chunks:", data_old.chunks)
print(f"Image size: ({SIZE},{SIZE})")
data_new = data_old.map_blocks(
shift_img,
chunks=data_old.chunks,
meta=np.array((), dtype=dtype),
)
# Write data_new to disk (triggers execution)
data_new.to_zarr(out_zarr)
# Clean up output folder
shutil.rmtree(out_zarr)
New, more flexible index-based approach to process the data
SIZE = 2000
# Function which increases value of each index in
# a SIZExSIZE image by one (=> pseudo-processing)
def shift_img(img):
return img.copy() + 1
def process_zarr(input_zarr):
out_zarr = f"out_{input_zarr}"
data_old = da.from_zarr(input_zarr)
data_new = da.empty(data_old.shape,
chunks=data_old.chunks,
dtype=data_old.dtype)
n_c, n_z, n_y, n_x = data_old.shape[:]
print(f"Input file: {input_zarr}")
print(f"Output file: {out_zarr}")
print("Array shape:", data_old.shape)
print("Array chunks:", data_old.chunks)
print(f"Image size: ({SIZE},{SIZE})")
for i_c in range(n_c):
for i_z in range(n_z):
for i_y in range(0, n_y - 1, SIZE):
for i_x in range(0, n_x - 1, SIZE):
# Need to do this in 2 steps to avoid a
# TypeError: Delayed objects of unspecified length have no len()
new_img = delayed_shift_img(data_old[i_c, i_z, i_y:i_y+SIZE, i_x:i_x+SIZE])
data_new[i_c, i_z, i_y:i_y+SIZE, i_x:i_x+SIZE] = da.from_delayed(new_img,
shape=(SIZE, SIZE),
dtype=data_old.dtype)
# Write data_new to disk (triggers execution)
data_new.to_zarr(out_zarr)
# Clean up output folder
shutil.rmtree(out_zarr)
Code to run the example based on different synthetic Zarr datasets & measure memory usage
input_zarrs = ['data_2_2_16000_16000.zarr', 'data_2_4_16000_16000.zarr', 'data_2_8_16000_16000.zarr', 'data_2_16_16000_16000.zarr']
for input_zarr in input_zarrs:
interval = 0.1
mem = memory_usage((process_zarr_mapblocks, (input_zarr,)), interval=interval)
#mem = memory_usage((process_zarr, (input_zarr,)), interval=interval)
time = np.arange(len(mem)) * interval
mem_file = "log_memory_" + input_zarr.split(".zarr")[0] + "_mapblocks.dat"
#mem_file = "log_memory_" + input_zarr.split(".zarr")[0] + "_.dat"
print(mem_file)
np.savetxt(mem_file, np.array((time, mem)).T)
What we observe
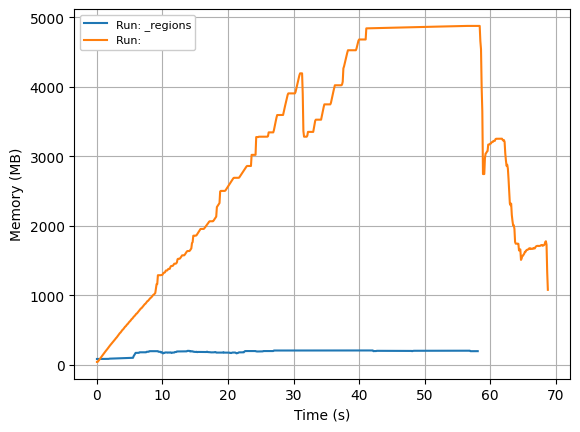
Using dask 2022.7.0 and python 3.9.12
For the mapblocks approach (full lines) to process the data, the runtime scales with the dataset size, but memory usage remains stable at < 1GB.
For the indexing approach (dotted lines), processing time also scales with the size of the dataset (a bit slower in this synthetic test case, but that seems to vary depending on test cases), but memory usage scales super-linear. While the small datasets (2, 2, 16k, 16k) & (2, 4, 16k, 16k) also use < 1 GB of memory, (2, 8, 16k, 16k) uses > 1 GB and (2, 16, 16k, 16k) peaks at almost 5 GB of memory usage.

Potential explanation:
When using our indexing approach, it appears that nothing is written to the zarr file until the end. That would explain why memory accumulates. But how do we change that? With the same to_zarr call in the mapblocks example, data is continuously written to the zarr file. Why isn’t this working in our delayed/indexing approach?
We describe our approach and the memory issue in further detail here: [ROI] Memory and running time for ROI-based illumination correction · Issue #131 · fractal-analytics-platform/fractal · GitHub
Tl;dr: How do we best process a Zarr file using dask in arbitrary chunks and how do we ensure that dask continuously writes to disk during the processing, not just at the end of processing?
Side note: I’ve started to test this in the 2022.08.0 version of dask in recent days and, while memory usage still scales super-linear (in a slightly different pattern), performance is about 30x slower with the indexing approach. I’ll open a separate dask issue for this performance slow-down between the dask releases. But for testing this, we were on 2022.07.0 only.